pacman::p_load(
tidyverse,
glue,
scales,
showtext,
ggtext,
shadowtext,
maps,
ggpattern,
ggrepel,
patchwork,
tidylog
)
font_add_google("Ubuntu", "Ubuntu", regular.wt = 400, bold.wt = 700)
showtext_auto()
showtext_opts(dpi = 300)
Tip
About the Data
Note
Warning
This dataset was removed from the TidyTuesday repository due to ethical reasons https://www.journals.uchicago.edu/doi/full/10.1086/693853
This week we are exploring lead levels in water samples collected in Flint, Michigan in 2015. The data comes from a paper by Loux and Gibson (2018) who advocate for using this data as a teaching example in introductory statistics courses.
The Flint lead data provide a compelling example for introducing students to simple univariate descriptive statistics. In addition, they provide examples for discussion
1 Initializing
1.1 Load libraries
1.2 Set theme
cool_gray0 <- "#323955"
cool_gray1 <- "#5a6695"
cool_gray2 <- "#7e89bb"
cool_gray3 <- "#a4aee2"
cool_gray4 <- "#cbd5ff"
cool_gray5 <- "#e7efff"
cool_red0 <- "#A31C44"
cool_red1 <- "#F01B5B"
cool_red2 <- "#F43E75"
cool_red3 <- "#E891AB"
cool_red4 <- "#FAC3D3"
cool_red5 <- "#FCE0E8"
theme_set(
theme_minimal() +
theme(
# axis.line.x.bottom = element_line(color = 'cool_gray0', linewidth = .3),
# axis.ticks.x= element_line(color = 'cool_gray0', linewidth = .3),
# axis.line.y.left = element_line(color = 'cool_gray0', linewidth = .3),
# axis.ticks.y= element_line(color = 'cool_gray0', linewidth = .3),
# # panel.grid = element_line(linewidth = .3, color = 'grey90'),
panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
axis.ticks.length = unit(-0.15, "cm"),
plot.background = element_blank(),
# plot.title.position = "plot",
plot.title = element_text(family = "Ubuntu", size = 14, face = 'bold'),
plot.caption = element_text(
size = 8,
color = cool_gray3,
margin = margin(20, 0, 0, 0),
hjust = 0
),
plot.subtitle = element_markdown(
size = 9,
lineheight = 1.15,
margin = margin(5, 0, 15, 0)
),
axis.title.x = element_markdown(
family = "Ubuntu",
hjust = .5,
size = 8,
color = cool_gray1
),
axis.title.y = element_markdown(
family = "Ubuntu",
hjust = .5,
size = 8,
color = cool_gray1
),
axis.text = element_text(
family = "Ubuntu",
hjust = .5,
size = 8,
color = cool_gray1
),
legend.position = "top",
text = element_text(family = "Ubuntu", color = cool_gray1),
# plot.margin = margin(25, 25, 25, 25)
)
)1.3 Load this week’s data
tuesdata <- tidytuesdayR::tt_load('2025-11-04')2 Quick Exploratory Data Analysis
2.1 Lead distribuition
tuesdata$flint_mdeq |>
ggplot(aes(x = lead)) +
geom_histogram(aes(fill = is.na(lead2)))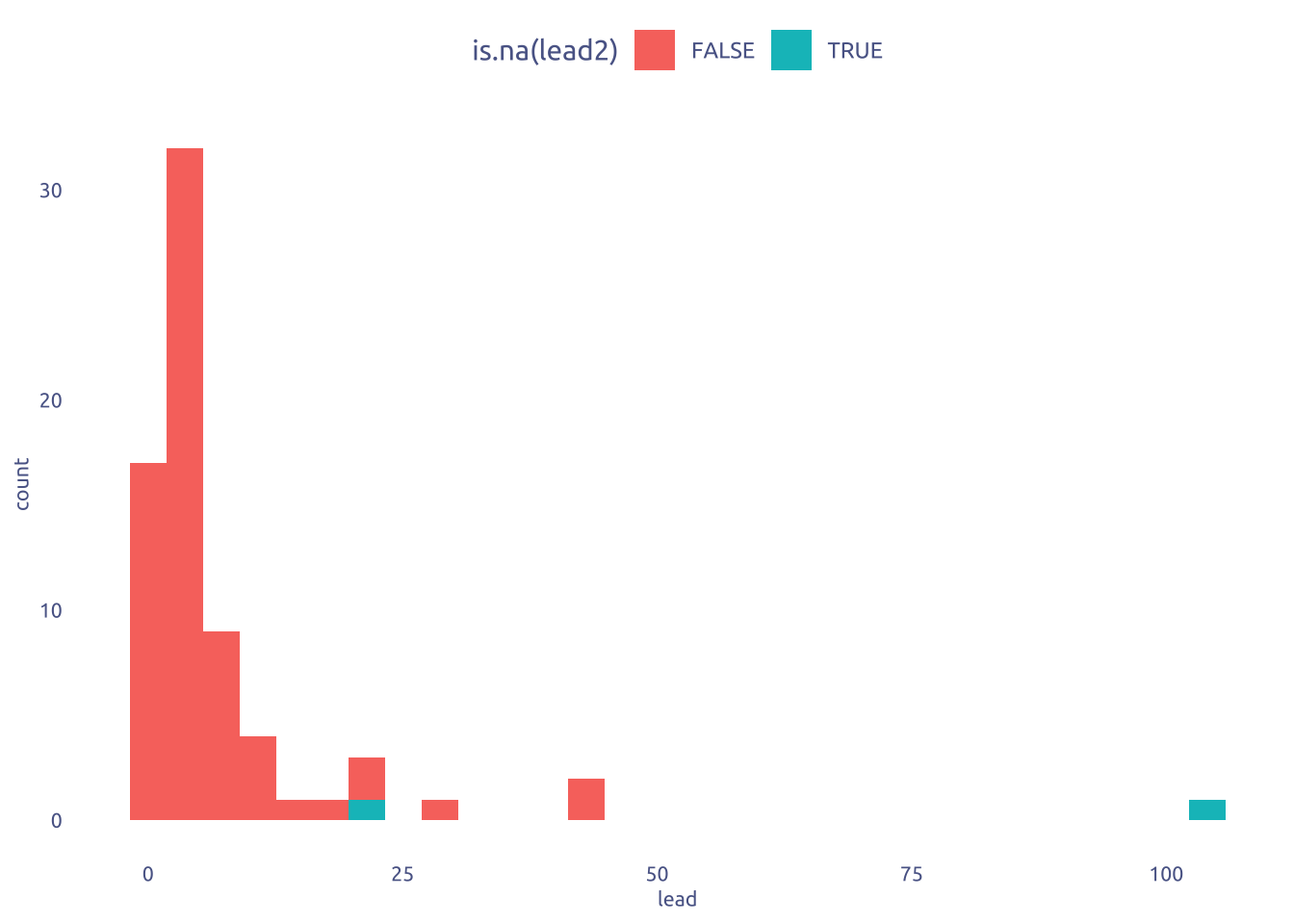
3 Transform Data for Plotting
data2plot <- tuesdata$flint_mdeq |>
transmute(
lead = lead,
removed = is.na(lead2),
notes = notes |> str_remove(".*: ") |> str_to_sentence()
)4 Time to plot!
4.1 Raw chart
data2plot |>
ggplot(aes(x = lead)) +
geom_histogram(aes(fill = removed)) +
theme_gray()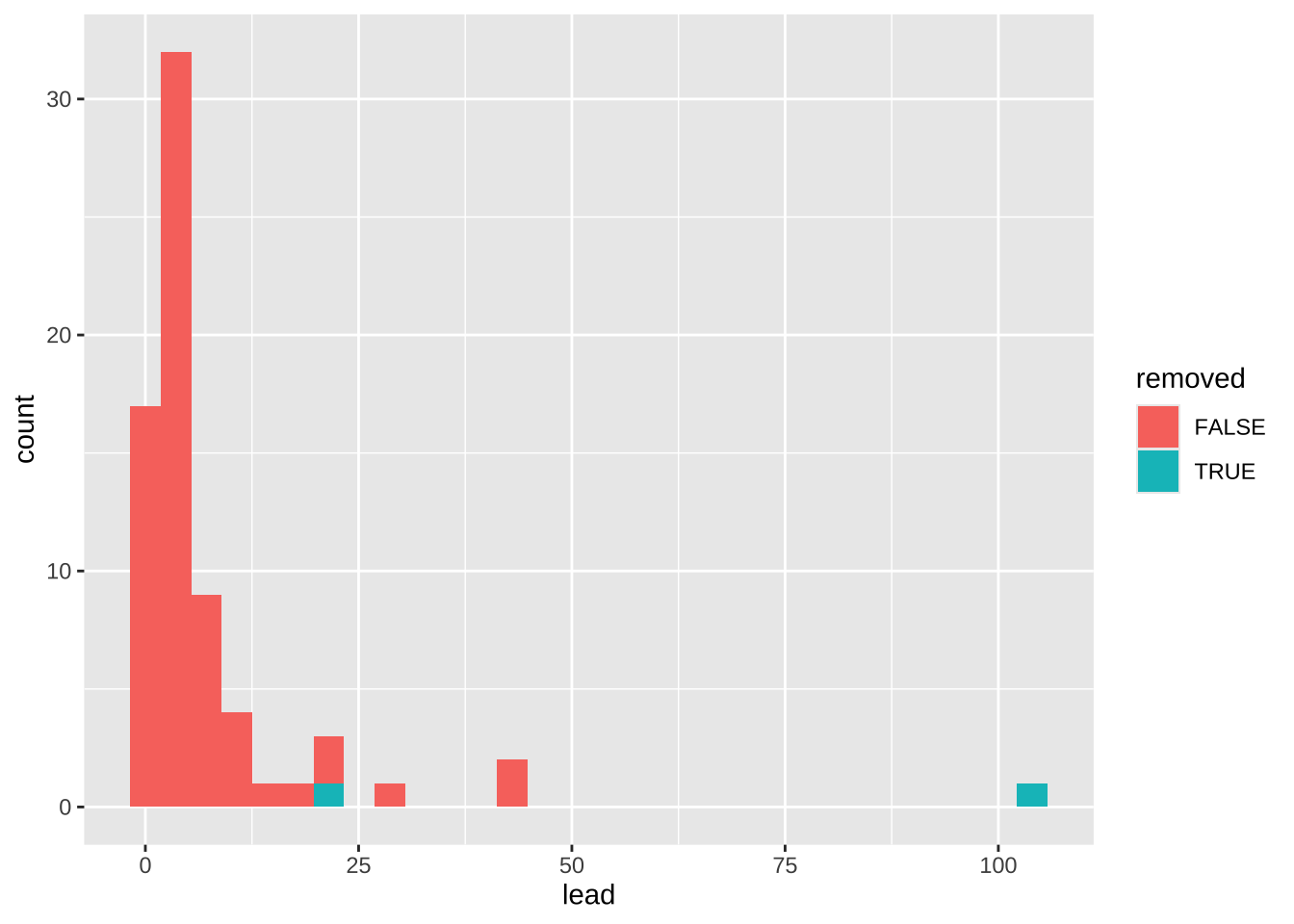
4.2 Final chart
quantile90_all <- data2plot |>
pull(lead) |>
quantile(0.9)
quantile90_correct <- data2plot |>
filter(!removed) |>
pull(lead) |>
quantile(0.9)
quantile90_EPA <- 15
#pseudorandom, smiley, maxout, frowney, minout, tukey, tukeyDense)
data2plot |>
ggplot(aes(x = lead)) +
ggbeeswarm::geom_beeswarm(
aes(fill = removed, y = "1"),
shape = 21,
color = cool_gray0,
cex = 3.5,
size = 2,
show.legend = FALSE
) +
annotate(
geom = "segment",
x = quantile90_all,
xend = quantile90_all,
y = -Inf,
yend = Inf,
linewidth = 0.5,
linetype = "dotted",
color = cool_red1
) +
annotate(
geom = "text",
x = quantile90_all,
y = 0.2,
vjust = 1.5,
hjust = 0,
label = "All samples",
angle = 90,
size = 3,
family = "Ubuntu",
color = cool_red1
) +
annotate(
geom = "segment",
x = quantile90_correct,
xend = quantile90_correct,
y = -Inf,
yend = Inf,
linewidth = 0.5,
linetype = "dotted",
color = cool_gray3
) +
annotate(
geom = "text",
x = quantile90_correct,
y = 0.2,
vjust = 1.5,
hjust = 0,
label = "Correct samples",
angle = 90,
size = 3,
family = "Ubuntu",
color = cool_gray2
) +
annotate(
geom = "segment",
x = quantile90_EPA,
xend = quantile90_EPA,
y = -Inf,
yend = Inf,
linewidth = 0.4,
color = cool_gray1
) +
annotate(
geom = "text",
x = quantile90_EPA,
y = 0.2,
vjust = 1.5,
hjust = 0,
label = "EPA threshold",
angle = 90,
size = 3,
family = "Ubuntu",
color = cool_gray1
) +
ggrepel::geom_text_repel(
data = data2plot |> filter(removed),
aes(label = notes),
hjust = 0,
vjust = -2.1,
size = 3,
family = "Ubuntu",
y = "1",
color = cool_red2,
min.segment.length = 99
) +
scale_fill_manual(values = c(cool_gray4, cool_red1)) +
theme(
axis.title.y = element_blank(),
axis.text.y = element_blank(),
panel.grid.major.x = element_line(
linewidth = .3,
color = cool_gray5
),
axis.title.x = element_markdown(
hjust = 0.045,
)
) +
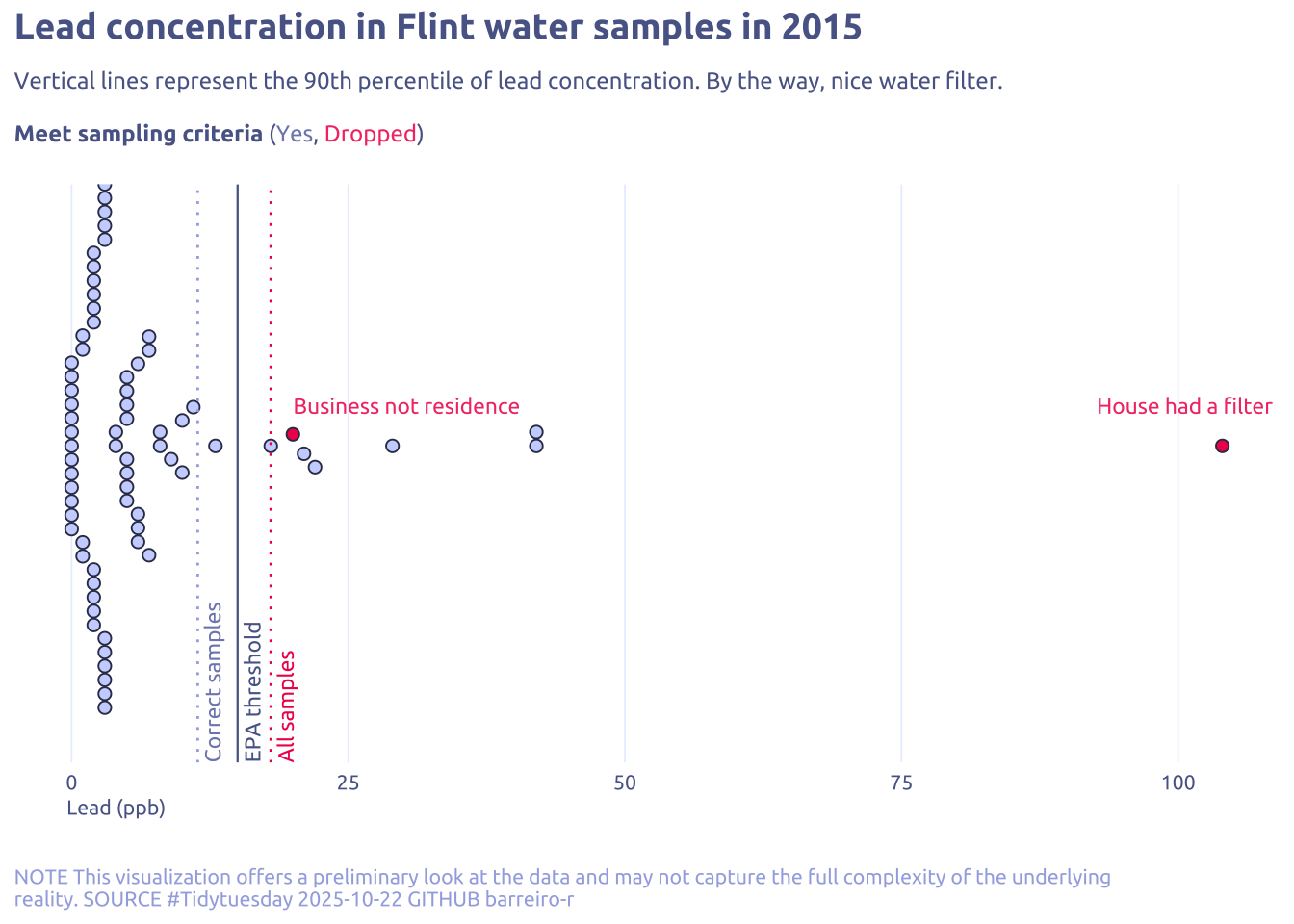
labs(
y = NULL,
title = "Lead concentration in Flint water samples in 2015",
subtitle = str_wrap(
'Vertical lines represent the 90th percentile of lead concentration. By the way, nice water filter.<br><br>**Meet sampling criteria** (<span style="color:#7e89bb">Yes</span>, <span style="color:#F43E75">Dropped</span>)',
width = 100,
),
caption = str_wrap(
"NOTE This visualization offers a preliminary look at the data and may not capture the full complexity of the underlying reality. SOURCE #Tidytuesday 2025-10-22 GITHUB barreiro-r",
width = 120,
),
x = "Lead (ppb)", fill = NULL
)